901829
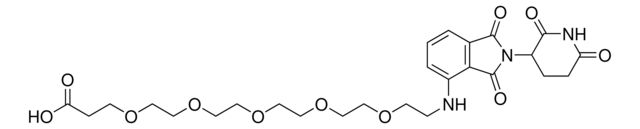
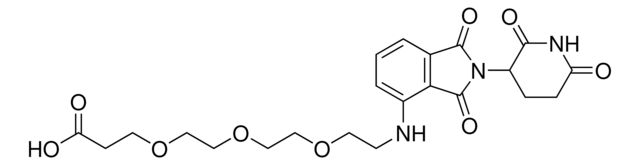
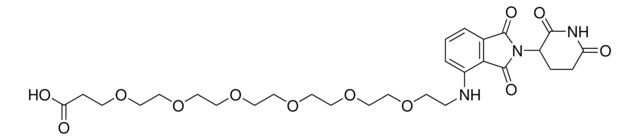
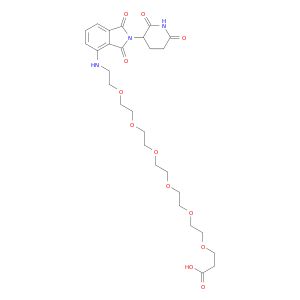
Pomalidomide-PEG6-CO2H
Manufacturer: Sigma Aldrich
CAS Number: 2225148-49-0
Synonym(S): 1-((2-(2,6-Dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)-3,6,9,12,15,18-hexaoxahenicosan-21-oic acid, Crosslinker−E3 Ligase ligand conjugate, Protein degrader building block for PROTAC® research, Template for synthesis of targeted protein degrader
Select a Size
| Pack Size | SKU | Availability | Price |
|---|---|---|---|
| 50 MG | 901829-50-MG | In Stock | ₹ 41,210.00 |
901829 - 50 MG
In Stock
Quantity
1
Base Price: ₹ 41,210.00
GST (18%): ₹ 7,417.80
Total Price: ₹ 48,627.80
ligand
pomalidomide
Assay
≥95%
form
liquid
reaction suitability
reactivity: amine reactivereagent type: ligand-linker conjugate
functional group
carboxylic acid
storage temp.
2-8°C
SMILES string
O=C(NC1=O)CCC1N(C2=O)C(C3=C2C=CC=C3NCCOCCOCCOCCOCCOCCOCCC(O)=O)=O
Other Options
| Image | Product Name | Manufacturer | Price Range | |
|---|---|---|---|---|
 | Pomalidomide-PEG6-COOH | Aaron Chemicals LLC | ₹ 8,989.00 - ₹ 27,234.00 | |
 | Pomalidomide-PEG6-C2-COOH | ChemScene | ₹ 10,235.00 - ₹ 91,136.00 | |
 | 2225148-49-0 | Pomalidomide-PEG6-COOH | A2B Chem | ₹ 10,235.00 - ₹ 30,349.00 |
Related Products
Description
- Application: Protein degrader builiding block Pomalidomide-PEG6-CO2H enables the synthesis of molecules for targeted protein degradation and PROTAC (proteolysis-targeting chimeras) technology. This conjugate contains a Cereblon (CRBN)-recruiting ligand and a PEGylated crosslinker with pendant carboxylic acid for reactivity with an amine on the target ligand. Because even slight alterations in ligands and crosslinkers can affect ternary complex formation between the target, E3 ligase, and PROTAC, many analogs are prepared to screen for optimal target degradation. When used with other protein degrader building blocks with a pendant carboxyl group, parallel synthesis can be used to more quickly generate PROTAC libraries that feature variation in crosslinker length, composition, and E3 ligase ligand.Automate your CRBN-PEG based PROTACs with Synple Automated Synthesis Platform (SYNPLE-SC002)
- Other Notes: TTechnology Spotlight: Degrader Building Blocks for Targeted Protein DegradationPortal: Building PROTAC® Degraders for Targeted Protein DegradationTargeted Protein Degradation by Small Molecules Small-Molecule PROTACS: New Approaches to Protein Degradations Targeted Protein Degradation: from Chemical Biology to Drug Discovery Impact of linker length on the activity of PROTACs
- Legal Information: PROTAC is a registered trademark of Arvinas Operations, Inc., and is used under license
SAFETY INFORMATION
WGK
WGK 3
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable